| 일 | 월 | 화 | 수 | 목 | 금 | 토 |
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | ||
| 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| 13 | 14 | 15 | 16 | 17 | 18 | 19 |
| 20 | 21 | 22 | 23 | 24 | 25 | 26 |
| 27 | 28 | 29 | 30 |
Tags
- flask
- Python
- ELASTIC
- high level client
- Kafka
- 900gle
- MySQL
- query
- TensorFlow
- zip 파일 암호화
- 차트
- Test
- token filter test
- springboot
- plugin
- matplotlib
- Mac
- license delete
- zip 암호화
- 파이썬
- docker
- aggregation
- Elasticsearch
- licence delete curl
- analyzer test
- sort
- API
- License
- aggs
- Java
Archives
- Today
- Total
개발잡부
[java] spark 본문
반응형
아래와 같은 에러가 난다면
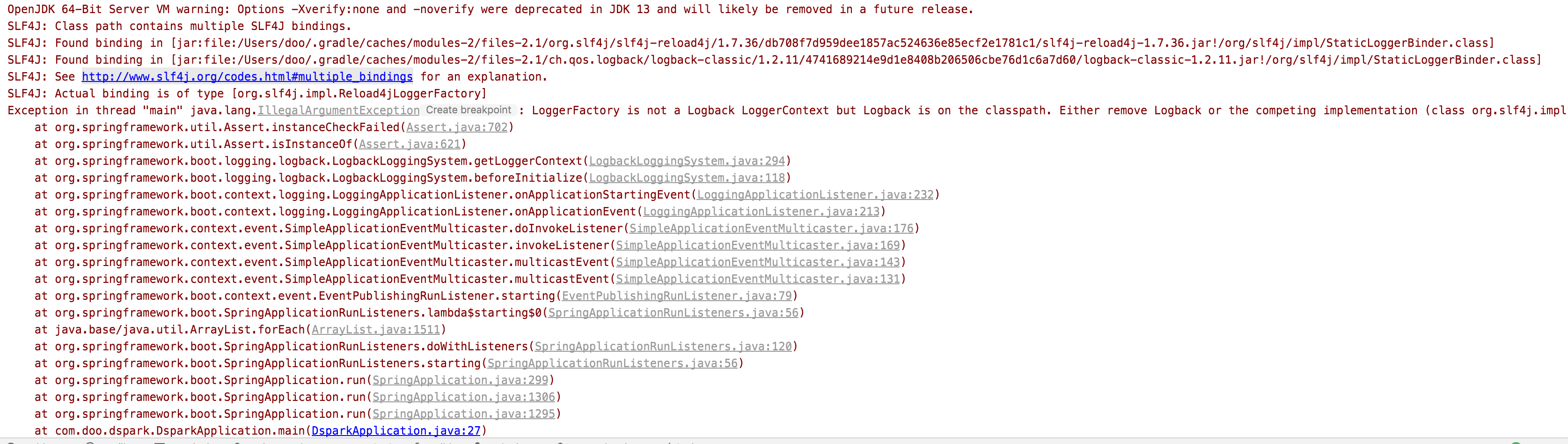
SLF4J: See http://www.slf4j.org/codes.html#multiple_bindings for an explanation.
Multiple bindings were found on the class path
configurations.all {
// 캐시하지 않음
resolutionStrategy.cacheChangingModulesFor 0, "seconds"
exclude group: "org.slf4j", module: "slf4j-log4j12"
}implementation("org.apache.spark:spark-sql_2.11:${sparkVersion}"){
exclude group: "org.slf4j", module: "slf4j-log4j12"
}
plugins {
id 'org.springframework.boot' version '2.7.0'
id 'io.spring.dependency-management' version '1.0.11.RELEASE'
id 'java'
}
group = 'com.doo'
version = '0.0.1-SNAPSHOT'
sourceCompatibility = '11'
configurations {
compileOnly {
extendsFrom annotationProcessor
}
}
repositories {
mavenCentral()
}
def sparkVersion = "3.0.1"
def elasticsearchVersion = "7.9.2"
configurations.all {
// 변하는 모듈(Changing Module)을 캐시하지 않음
resolutionStrategy.cacheChangingModulesFor 0, "seconds"
exclude group: "org.slf4j", module: "slf4j-log4j12"
exclude group: 'org.codehaus.janino', module: 'commons-compiler'
}
dependencies {
implementation 'org.springframework.boot:spring-boot-starter-actuator'
implementation 'org.springframework.boot:spring-boot-starter-web'
compileOnly 'org.projectlombok:lombok'
annotationProcessor 'org.projectlombok:lombok'
// spark
implementation("org.apache.spark:spark-core_2.12:${sparkVersion}")
implementation("org.apache.spark:spark-mllib_2.12:${sparkVersion}")
implementation("org.apache.spark:spark-mllib-local_2.12:${sparkVersion}")
implementation("org.apache.spark:spark-sql_2.12:${sparkVersion}")
implementation("org.apache.spark:spark-catalyst_2.12:${sparkVersion}")
implementation("org.elasticsearch:elasticsearch-spark-20_2.11:${elasticsearchVersion}")
implementation("mysql:mysql-connector-java:5.1.45")
// https://mvnrepository.com/artifact/org.codehaus.janino/commons-compiler
implementation group: 'org.codehaus.janino', name: 'commons-compiler', version: '3.0.1'
testImplementation 'org.springframework.boot:spring-boot-starter-test'
}
tasks.named('test') {
useJUnitPlatform()
}
package com.doo.dspark;
import lombok.Data;
import org.apache.spark.ml.classification.RandomForestClassificationModel;
import org.apache.spark.ml.classification.RandomForestClassifier;
import org.apache.spark.ml.evaluation.MulticlassClassificationEvaluator;
import org.apache.spark.ml.feature.StringIndexer;
import org.apache.spark.ml.feature.VectorAssembler;
import org.apache.spark.sql.Column;
import org.apache.spark.sql.Dataset;
import org.apache.spark.sql.Row;
import org.apache.spark.sql.SparkSession;
import org.apache.spark.sql.types.DataTypes;
import org.springframework.boot.CommandLineRunner;
import org.springframework.boot.SpringApplication;
import org.springframework.boot.autoconfigure.SpringBootApplication;
import org.springframework.context.annotation.Bean;
import org.springframework.context.annotation.Profile;
import static org.apache.spark.sql.functions.*;
@SpringBootApplication
public class DsparkApplication {
public static void main(String[] args) {
SpringApplication.run(DsparkApplication.class, args);
}
@Profile("!test")
@Bean
public CommandLineRunner runme() {
return args -> {
runSparkML();
};
}
public void runSparkML(){
SparkSession ss = SparkSession.builder()
.appName("TitanicSurvive")
.config("spark.eventLog.enabled", "false")
.master("local[1]")
.getOrCreate();
ss.sparkContext().setLogLevel("ERROR");
Dataset<Row> rows = ss.read().option("header", true).csv("/Users/doo/project/dspark/train.csv");
//Selection of useful columns (features)
System.out.println("==========Select useful colmns and casting========");
rows = rows.select(
col("Survived").cast(DataTypes.IntegerType),
col("Pclass").cast(DataTypes.DoubleType),
col("Sex"),
col("Age").cast(DataTypes.DoubleType),
col("Fare").cast(DataTypes.DoubleType),
col("Embarked")
);
System.out.println("Dataframe:");
rows.show();
System.out.println("Scheme:");
rows.printSchema();
//How the deta looks like
System.out.println("==========Preliminary Statistic==========");
System.out.println("Total passenger count: " + rows.count());
Dataset<Row> describe = rows.describe();
describe.show();
//Data cleansing -- Study if Null is a problem (Yes, in our case)
System.out.println("==========How many rows are with null value?==========");
String colNames[] = rows.columns();
Dataset<Row> summary = null;
for(int i = 0; i < colNames.length; i++){
String thisColName = colNames[i];
Dataset<Row> numNullinCol = rows.filter(col(thisColName).isNull()).select(count("*").as("NullOf"+thisColName));
/*
To reader: Please suggest better way to do counting of null values in each columns using Spark Java API
*/
if(summary==null){
summary=numNullinCol;
continue;
}
summary = summary.join(numNullinCol);
}
summary.show();
//Data cleansing - Remove Null Values
System.out.println("==========Eliminate Row with null values==========");
Column dropCondition = null;
for(int i = 0; i < colNames.length; i++){
Column filterCol = col(colNames[i]).isNotNull();
if(dropCondition==null) {
dropCondition = filterCol;
continue;
}
dropCondition = dropCondition.and(filterCol);
}
System.out.println("Filter condition: " + dropCondition);
Dataset<Row> nonNullPassengers = rows.filter(dropCondition);
System.out.println("Remain number of non-null passenger: " + nonNullPassengers.count());
nonNullPassengers.show();
//Feature Engineering -- Prepare
System.out.println("========Feature preparation - Index String to numbers for Spark MLib========");
StringIndexer sexIndexer = new StringIndexer().setInputCol("Sex").setOutputCol("Gender").setHandleInvalid("keep");
StringIndexer embarkedIndexer = new StringIndexer().setInputCol("Embarked").setOutputCol("Boarded").setHandleInvalid("keep");
Dataset<Row> genderPassengers = sexIndexer.fit(nonNullPassengers).transform(nonNullPassengers);
Dataset<Row> boardedGenderPassengers = embarkedIndexer.fit(genderPassengers).transform(genderPassengers);
boardedGenderPassengers = boardedGenderPassengers.drop("Sex", "Embarked");
boardedGenderPassengers.printSchema();
boardedGenderPassengers.show();
System.out.println("The column Gender and Boarded have been indexed to map to numbers");
//Feature Engineering -- Prepare
System.out.println("========Feature engineering ========");
String featureCol[] = {
"Pclass",
"Age",
"Fare",
"Gender",
"Boarded"
};
//Be aware of the mistake to pass the label(ie observed result) "Survived" column, model will be 100% correct if so...too good to be true
VectorAssembler vAssembler = new VectorAssembler();
vAssembler.setInputCols(featureCol).setOutputCol("features");
Dataset<Row> featureReadyDF = vAssembler.transform(boardedGenderPassengers);
featureReadyDF.printSchema();
featureReadyDF.show();
//Modeling -- Training data and Testing data
System.out.println("========Modeling - Use same data set as both training and testing -- NOT best practice ========");
//Actually, we should use the "seen" data "train.csv" to train model and "unseen" "test.csv" for validation.
//As we don't want to download the "test.csv" for the test part, we just split existing "train.csv" -> "80% train, 20% test" (pretend we haven't see 20% test data)
//Because of simpliticy of this example, bias/look-ahead didn't happen;
Dataset<Row>[] bothTrainTestDFs = featureReadyDF.randomSplit(new double[]{0.8d,0.2d});
Dataset<Row> trainDF = bothTrainTestDFs[0];
Dataset<Row> testDF = bothTrainTestDFs[1];
System.out.println("===Training set===");
trainDF.printSchema();
trainDF.show();
System.out.println("Total record: " + trainDF.count());
System.out.println("===Testing set===");
testDF.printSchema();
testDF.show();
System.out.println("Total record: " + testDF.count());
//Modeling -- Machine learning (train an estimator)
System.out.println("========Modeling - Building a model with an estimator ========");
RandomForestClassifier estimator = new RandomForestClassifier()
.setLabelCol("Survived")
.setFeaturesCol("features")
.setMaxDepth(5);
RandomForestClassificationModel model = estimator.fit(trainDF);
//we ask the Estimator to learn from training data
//Now we have the learnt `model` to be used with test data and see how good this model is
//Modeling -- Prediction with existing data (Given some features, what is the prediction?)
System.out.println("========Modeling - Use an estimator to predict if the passenger survived==========");
Dataset<Row> predictions = model.transform(testDF);
System.out.println("Here is the predictions:");
predictions.printSchema();
predictions.show();
//Modeling -- How good is our predictions?
System.out.println("========Modeling - Check how good is the model we've built ==========");
MulticlassClassificationEvaluator evaluator = new MulticlassClassificationEvaluator()
.setLabelCol("Survived")
.setPredictionCol("prediction");
double accuracy = evaluator.evaluate(predictions);
System.out.println("Accuracy: " + accuracy);
}
//You might be tempted to use Dataset<Passenger> but since we are going to add/delete/move/transfer column around, this is not used
@Data
public static class Passenger{
//Integer PassengerId;
Integer Survived;
Double Pclass;
//String Name;
String Sex;
Double Age;
//Integer SibSp;
//Integer Parch;
//Integer Ticket;
Double Fare;
//String Cabin;
String Embarked;
}
}
반응형
'etc.' 카테고리의 다른 글
| Spark install (0) | 2022.06.16 |
|---|---|
| Process 'command '/Library/Java/JavaVirtualMachines/adoptopenjdk-15.jdk/Contents/Home/bin/java'' finished with non-zero exit value 1 (0) | 2022.06.05 |
| [Sourcetree] 푸시하기 (0) | 2022.04.05 |
| MacBook Host 수정 (0) | 2022.03.14 |
| 가족관계증명서 발급하기 - 법원 (0) | 2022.01.19 |
Comments


